C20orf141
| C20orf141 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | C20orf141, dJ860F19.4, chromosome 20 open reading frame 141 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1922906; HomoloGene: 50959; GeneCards: C20orf141; OMA:C20orf141 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Chromosome 20 open reading frame 141 (c20orf141) is a protein encoded by the c20orf141 gene in Humans (Homo sapiens).[5] Human c20orf141 is located on the short arm of Chromosome 20 in region 1 and band 3.[6] It is a single-pass membrane protein predicted to be localized in the membrane, and is a member of the uncharacterized DUF5562 (PF17717) protein superfamily.[7]
Gene
Human c20orf141 spans 5,136 base pairs, from positions 2,795,614 to 2,800,930 on the plus strand of Chromosome 20 (20p13). [8] It sits downstream of a carboxypeptidase-encoding gene and upstream of a vacuolar protein sorting gene.[5] C20orf141 sits upstream of PC-Esterase Domain Containing 1A (PCED1A), Receptor-Type Tyrosine-Protein Phosphatase Alpha (PTPRA), Vacuolar Protein Sorting Protein 16 (VPS16).[6] C20orf141 is also downstream of Carboxypeptidase X1 (CPXM1) and overlaps with Transmembrane Protein 239 (TMEM239).[6]
C20orf141 is also known under the aliases MGC26144 and LOC128653. [8]

RNA transcript
Human c20orf141 has two mRNA transcript variants, differing in 5' and 3' untranslated regions (UTRs).[8] mRNA transcript variant 1 contains 2 exons and 1 intron, while mRNA transcript variant 2 contains 3 exons and 2 introns.[8] Both mRNA variants encode the same protein isoform.[8]
Protein
Human c20orf141 is predicted to be a single-pass membrane protein.[9] The protein encoded by the gene c20orf141 has 165 amino acids, a molecular weight of ~17.4 kDa, and an isoelectric point of ~7.8.[9][10][11] There is only one human isoform of c20orf141.[9] C20orf141 is leucine-rich compared to other human proteins but poor in asparagine, tyrosine, and lysine.[12] DTU Health predicts six different phosphorylation sites at Ser31, Ser66, Ser50, Thr67, Ser104, and Ser129.[13]
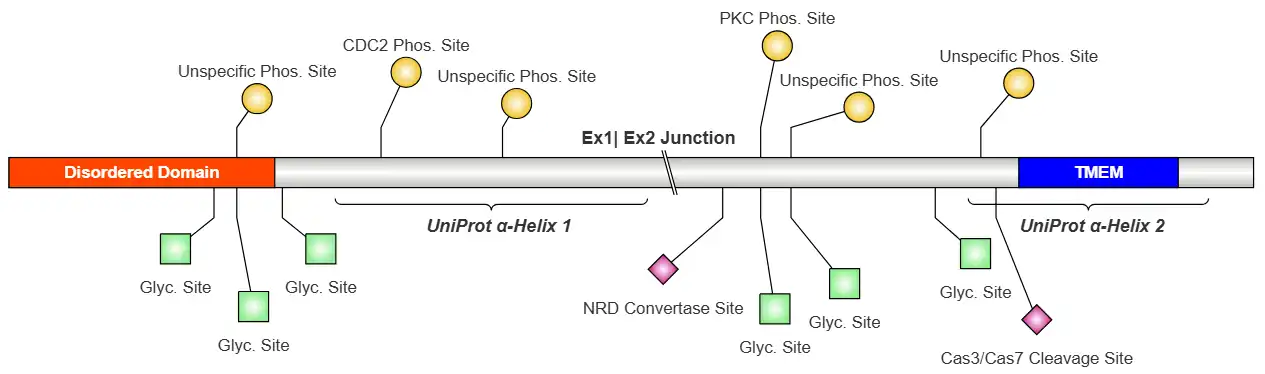
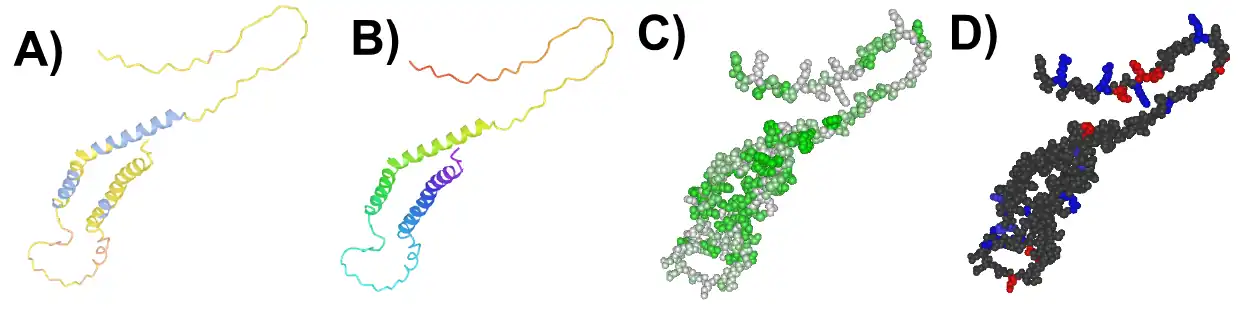
Human c20orf141 has a disordered region and transmembrane domain.[9] However, human c20orf141 has two predicted alpha helices in concentrated clusters of hydrophobic amino acids. [14][15][16]
Localization
Rabbit c20orf141 protein is localized in the plasma membrane of cells, as identified by an immunocytochemistry analysis of the GAMG cell line.[17] Human c20orf141 is predicted to be subcellularly localized in membrane-bound organelles, specifically the endoplasmic reticulum according to PSORT II. [18]
Conceptual translation
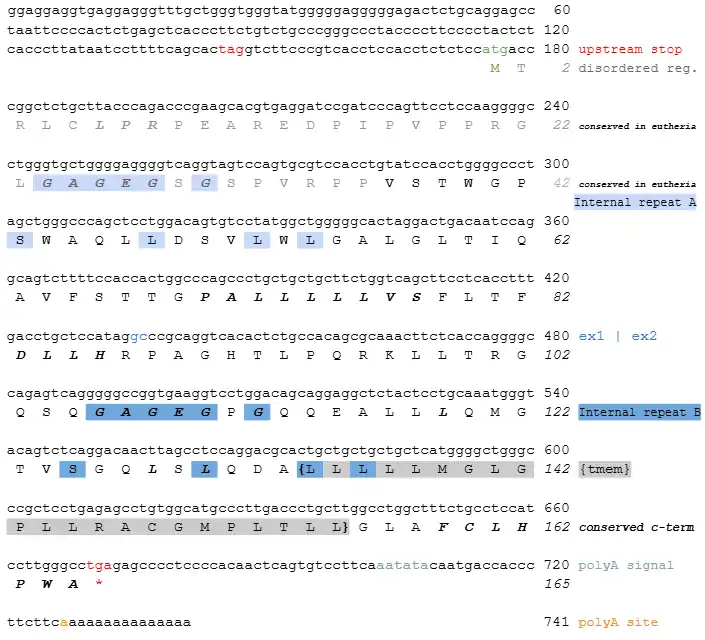
Protein-protein interactions
Human c20orf141 is an interaction partner with Apolipoprotein E (APOE), Ataxin-1 (ATXN1), Huntingtin (HTT), and Junctophilin-3 (JPH3), which are involved in neuronal development and implicated in neurological disorders like Bipolar Disorder, Huntington's Disease, and Alzheimer's.[19] One study investigating laminopathies found no evidence that the protein encoded by human c20orf141 binds to or interacts with lamin A, a structural integrity protein. [20] In addition to neurological-associated proteins, human c20orf141 binds to major inflammatory and immune response regulators like ENDOD1, IL17F, and ZDHHC17. [21] Human 20orf141 also engages in protein-protein interactions with CCDC12, FUNDC2, and ACTA2, which are involved in blood cell maintenance and survival. [21]
Expression
Tissues
Human c20orf141 is expressed highly in the placenta, testis, and epididymis.[8][22] Moderate levels of human c20orf141 expression are also seen in the adrenal gland, fetal brain, heart, fetal kidney, skeletal muscle, ovary, and skin tissues.[22] Expression is observed to be incredibly low in the colon and retina tissue.[22]
Conditions
Human c20orf141 expression in intestinal cells is relatively low when linoleic acid or a mixture of trans-10 and cis-12 conjugated linoleic acid (CLA) is present.[23] However, when both cis-9 and trans-11 CLA isomers, associated with anti-inflammatory responses, are present, c20orf141 expression drops significantly and becomes in the bottom 25th percentile of gene expression.[23][24]
c20orf141 baseline expression is relatively low in tubular cells.[25] Expression is also unchanged in the presence of only C-peptide or a mixture of TGF-β1 and C-peptide.[25] However, c20orf141 expression drops to very low levels when in the presence of only TGF-β1.[25]
At baseline levels in CUTLL1 cells, expression is low to moderate (just above the 50th percentile of overall expression).[26] The transduction of MigR1 and DN-MAML plasmid vectors to activate the Notch cell-communication pathway results in lower expression of c20orf141.[26]
eQTLs
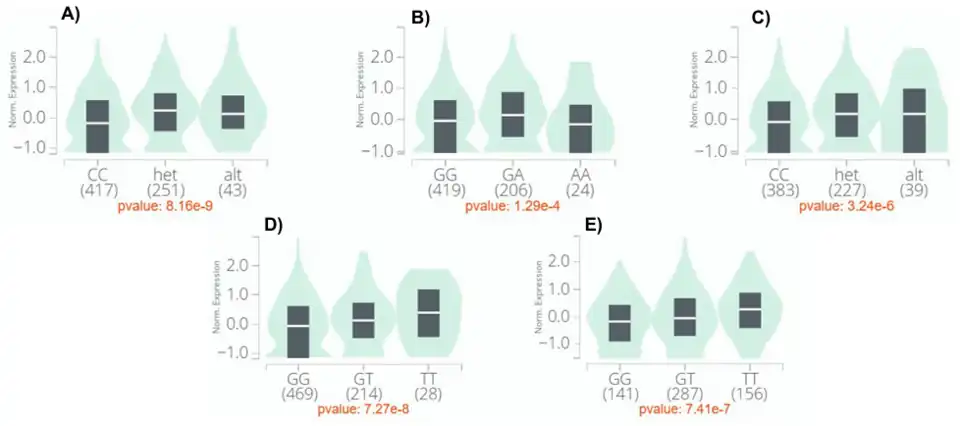
All known expression quantitative trait loci (eQTLs) of c20orf141 are found downstream of the 3' untranslated region (UTR).[27] In subcutaneous and visceral (omentum) adipose tissue, homozygous genotypes have lower expression, while alternative allele genotypes have the highest.[27] Similarly, the homozygous genotype in fibroblast cells has the lowest expression. [27]
Clinical significance
Human c20orf141 is upregulated in clear cell renal cell carcinoma (ccRCC).[28] Human c20orf141 is downregulated in the sigmoid colon's mucosa (inner lining) in irritable bowel syndrome. [29]
Evolution
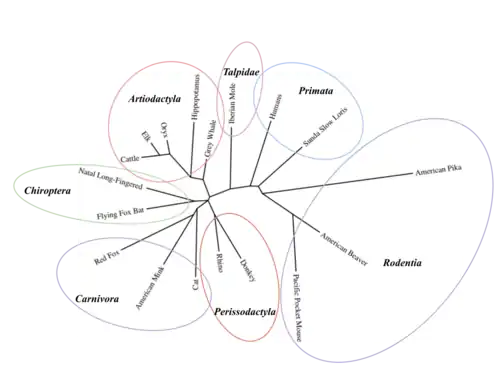
C20orf141 orthologs are found only in mammals, specifically in the infraclasses metatheria and eutheria.[8] Orthologs of c20orf141 are specifically found in Primata, Rodentia, Artiodactyla, Perissodactyla, Talpidae, Chiroptera, and Carnivora.[10] Within eutheria, c20orf141 orthlogs are only found in opposums.[10] C20orf141 has no known paralogs.[10]
C20orf141 is likely to have emerged 160 million years ago (MYA), as the most distant ortholog to humans is found in opossums.[30]
The protein sequence of c20orf141 has two predicted internal repeats.[9][31] However, only its second internal repeat at Met1-Gly55 and Asp73-Leu137 is conserved among orthologous sequences.[9]
| Genus | Common Name | Taxon | Date of Divergence (MYA) | Accession Number | Sequence Number (AA) | Sequence Identity % | Sequence Similarity % |
|---|---|---|---|---|---|---|---|
| Homo sapiens | Humans | Primates | 0.0 | NP_542777.1 | 165 | 100.0 | 100.0 |
| Pan troglodytes | Chimpanzee | Primates | 6.4 | XP_001159993.1 | 165 | 98.2 | 98.8 |
| Nycticebus coucang | Sunda slow loris | Primates | 74.0 | XP_053430470.1 | 165 | 77.2 | 82.0 |
| Urocitellus parryii | Arctic squirrel | Rodentia | 87.0 | XP_026265484.1 | 167 | 73.7 | 80.8 |
| Castor canadensis | American beaver | Rodentia | 87.0 | XP_020029419.2 | 261 | 71.4 | 76.8 |
| Perognathus longimembris | Pocket mouse | Rodentia | 87.0 | XP_048205328.1 | 164 | 69.2 | 74.8 |
| Ochotona princeps | American pika | Rodentia | 87.0 | XP_004585728.2 | 165 | 69.5 | 76.7 |
| Mus musculus | House mice | Rodentia | 87.0 | NP_001156955.1 | 131 | 64.6 | 73.2 |
| Talpa occidentalis | Iberian mole | Eulipotyphla | 94.0 | XP_037377215.1 | 167 | 72.7 | 76.4 |
| Pteropus vampyrus | Flying fox bat | Chiroptera | 94.0 | XP_011367784.1 | 167 | 73.6 | 77.8 |
| Miniopterus natalensis | Natal bat | Chiroptera | 94.0 | XP_016071282.1 | 173 | 68.8 | 74.6 |
| Felis catus | Domestic cat | Carnivora | 94.0 | XP_003983796.2 | 167 | 71.1 | 78.3 |
| Neogale vison | American mink | Carnivora | 94.0 | XP_044118321.1 | 167 | 70.1 | 76.0 |
| Vulpes vulpes | Red fox | Carnivora | 94.0 | XP_025868051.1 | 168 | 69.0 | 75.6 |
| Eschrichtius robustus | Grey whale | Whippomorpha | 94.0 | XP_068381043.1 | 165 | 75.3 | 80.1 |
| Hippopotamus amphibius | Hippo | Whippomorpha | 94.0 | XP_057557119.1 | 167 | 72.5 | 77.3 |
| Lipotes vexillifer | Baiji Dolphin | Whippomorpha | 94.0 | XP_007465451.1 | 172 | 71.5 | 77.9 |
| Equus asinus | Donkey | Perissodactyla | 94.0 | XP_014717342.1 | 167 | 75.5 | 78.4 |
| Cervus canadensis | Elk | Cervidae | 94.0 | XP_043336499.1 | 163 | 75.0 | 80.5 |
| Oryx dammah | Scimitar oryx | Bovidae | 94.0 | XP_040103245.1 | 165 | 74.7 | 80.7 |
| Diceros bicornis minor | Black Rhino | Perissodactyla | 94.0 | KAF5924871.1 | 164 | 74.2 | 74.1 |
| Bos taurus | Domestic cattle | Bovidae | 94.0 | XP_002692279.2 | 165 | 73.5 | 79.5 |
| Gracilinanus agilis | Mouse Opossum | Marsupialia | 160.0 | XP_044537729.1 | 188 | 38.2 | 50.9 |
| Monodelphis domestica | Gray Opossum | Marsupialia | 160.0 | XP_007497002.2 | 239 | 36.6 | 50.0 |
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000258713 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000027409 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "AceView: geneid:C20orf141, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView". www.ncbi.nlm.nih.gov. Retrieved 2025-07-29.
- ^ a b c "C20orf141 chromosome 20 open reading frame 141 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2025-08-03.
- ^ "NCBI c20orf141 Transcript Variant 1". NCBI c20orf141 Transcript Variant 1. 2025.
- ^ a b c d e f g "C20orf141 chromosome 20 open reading frame 141 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2025-07-29.
- ^ a b c d e f "uncharacterized protein C20orf141 [Homo sapiens] - Protein - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2025-07-29.
- ^ a b c d GeneCards Human Gene Database. "C20orf141 Gene - GeneCards | CT141 Protein | CT141 Antibody". www.genecards.org. Archived from the original on 2022-12-02. Retrieved 2025-07-29.
- ^ "Expasy - Compute pI/Mw tool". web.expasy.org. Retrieved 2025-08-03.
- ^ "SAPS Sequence Dispatcher". www.ebi.ac.uk. Retrieved 2025-07-29.
- ^ "NetPhos 3.1 - DTU Health Tech - Bioinformatic Services". services.healthtech.dtu.dk. Retrieved 2025-08-03.
- ^ a b "UniProt". UniProt. Retrieved 2025-07-29.
- ^ a b "iCn3D: Web-based 3D Structure Viewer". www.ncbi.nlm.nih.gov. Retrieved 2025-07-29.
- ^ a b "ProP 1.0 - DTU Health Tech - Bioinformatic Services". services.healthtech.dtu.dk. Retrieved 2025-07-29.
- ^ "Anti-C20orf141 Antibodies | Invitrogen". www.thermofisher.com. Retrieved 2025-07-29.
- ^ "PSORT II Prediction". psort.hgc.jp. Retrieved 2025-07-29.
- ^ Haenig C, Atias N, Taylor AK, Mazza A, Schaefer MH, Russ J, et al. (2020-08-18). "Interactome Mapping Provides a Network of Neurodegenerative Disease Proteins and Uncovers Widespread Protein Aggregation in Affected Brains". Cell Reports. 32 (7) 108050: 108050. doi:10.1016/j.celrep.2020.108050. ISSN 2211-1247. PMID 32814053.
- ^ Dittmer TA, Sahni N, Kubben N, Hill DE, Vidal M, Burgess RC, et al. (2014). "Systematic identification of pathological lamin A interactors". Molecular Biology of the Cell. 25 (9): 1493–1510. doi:10.1091/mbc.e14-02-0733. ISSN 1059-1524. PMC 4004598. PMID 24623722.
- ^ a b "IntAct Portal". www.ebi.ac.uk. Retrieved 2025-07-29.
- ^ a b c Dezso Z, Nikolsky Y, Sviridov E, Shi W, Serebriyskaya T, Dosymbekov D, et al. (2008). "A comprehensive functional analysis of tissue specificity of human gene expression". BMC Biology. 6 49: 49. doi:10.1186/1741-7007-6-49. ISSN 1741-7007. PMC 2645369. PMID 19014478.
- ^ a b Murphy EF, Hooiveld GJ, Muller M, Calogero RA, Cashman KD (2007). "Conjugated linoleic acid alters global gene expression in human intestinal-like Caco-2 cells in an isomer-specific manner". The Journal of Nutrition. 137 (11): 2359–2365. doi:10.1093/jn/137.11.2359. ISSN 0022-3166. PMID 17951470.
- ^ Wang T, Lee HG (2015-04-16). "Advances in Research on cis-9, trans-11 Conjugated Linoleic Acid: A Major Functional Conjugated Linoleic Acid Isomer". Critical Reviews in Food Science and Nutrition. 55 (5): 720–731. doi:10.1080/10408398.2012.674071. ISSN 1040-8398. PMID 24915361.
- ^ a b c Hills CE, Al-Rasheed N, Al-Rasheed N, Willars GB, Brunskill NJ (2008). "C-peptide reverses TGF-beta1-induced changes in renal proximal tubular cells: implications for treatment of diabetic nephropathy". American Journal of Physiology. Renal Physiology. 296 (3): F614 – F621. doi:10.1152/ajprenal.90500.2008. ISSN 1931-857X. PMC 2660188. PMID 19091788.
- ^ a b Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, et al. (2011-09-06). "Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells". Proceedings of the National Academy of Sciences of the United States of America. 108 (36): 14908–14913. Bibcode:2011PNAS..10814908W. doi:10.1073/pnas.1109023108. ISSN 1091-6490. PMC 3169118. PMID 21737748.
- ^ a b c d "GTEx Portal". www.gtexportal.org. Retrieved 2025-07-29.
- ^ Al Sharie AH, Al Masoud EB, Jadallah RK, Alzghoul SM, Darweesh RF, Al-Bataineh R, et al. (2024-09-27). "Transcriptome analysis revealed a novel nine-gene prognostic risk score of clear cell renal cell carcinoma". Medicine. 103 (39): e39678. doi:10.1097/MD.0000000000039678. PMC 11441924. PMID 39331921.
- ^ Videlock EJ, Mahurkar-Joshi S, Hoffman JM, Iliopoulos D, Pothoulakis C, Mayer EA, et al. (2018). "Sigmoid colon mucosal gene expression supports alterations of neuronal signaling in irritable bowel syndrome with constipation". American Journal of Physiology. Gastrointestinal and Liver Physiology. 315 (1): G140 – G157. doi:10.1152/ajpgi.00288.2017. ISSN 0193-1857. PMC 6109711. PMID 29565640.
- ^ "TimeTree :: The Timescale of Life". timetree.org. Retrieved 2025-07-29.
- ^ "Dot Analysis - Internal Repeats". www.ebi.ac.uk. Retrieved 2025-08-03.