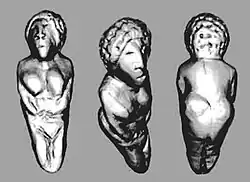
Ancient North Eurasian
Mal'ta–Buret' culture ivory figurines (c. 24,000 BP-c. 15,000 BP). Some of the figurines wear hooded overalls with decorative stripes.[1][2] |
In archaeogenetics, the term Ancient North Eurasian (ANE) refers to an ancestral component that represents the lineage of the people of the Mal'ta–Buret' culture (c. 24,000 BP) and populations closely related to them, such as the Upper Paleolithic individuals from Afontova Gora in Siberia.[6][7] Genetic studies also revealed that the ANE are closely related to the remains of the preceding Yana culture (c. 32,000 BP), which were dubbed as Ancient North Siberians (ANS), and which either are directly ancestral to the ANE, or both being closely related sister lineages.[8]
The ANE/ANS lineages both derive their ancestry from an admixture event between 'Ancient West Eurasians' (best represented by Upper Paleolithic Europeans such as Kostenki-14, c. 38,000 BP) and 'Ancient East Eurasians' (best represented by the Tianyuan man, c. 39,000 BP) during the Upper Paleolithic period.[9][8][10][11]
Around 20,000 to 25,000 years ago, a branch of Ancient North Eurasian people mixed with Ancient East Asians, which led to the emergence of Ancestral Native American, Ancient Beringian and Ancient Paleo-Siberian populations. It is unknown exactly where this population admixture took place, and two opposing theories have put forth different migratory scenarios that united the Ancient North Eurasians with ancient East Asian populations.[12]
Later, ANE populations migrated westward into Europe and admixed with European Western hunter-gatherer (WHG)-related groups to form the Eastern hunter-gatherer (EHG) group, which later admixed with Caucasus hunter-gatherers to form the Western Steppe Herder group, which became widely dispersed across Eurasia during the Bronze Age.[13]
ANE ancestry has spread throughout Eurasia and the Americas in various migrations since the Upper Paleolithic.[14] Significant ANE ancestry can be found in Native Americans, as well as in Europe, South Asia, Central Asia, and Siberia. It has been suggested that their mythology may have featured narratives shared by both Indo-European and some Native American cultures, such as the existence of a metaphysical world tree and a dog which guards the path to the afterlife.[15]
Genetic studies
Definition
_with_tomb_artifacts%252C_Hermitage_Museum%252C_Saint-Petersburg.jpg)
The ANE lineage, also known as Paleolithic Siberians, is defined by association with the "Mal'ta boy" (MA-1), the remains of an individual who lived during the Last Glacial Maximum, 24,000 years ago in central Siberia, discovered in the 1920s. Together with the Yana Rhinoceros Horn Site samples, and Afontova Gora individuals, they are collectively referred to as 'Ancient North Siberians', although 'Ancient North Eurasian' is also used as a collective nname for both MA-1 and Yana remains.[5][4][10][8]
Ancient North Eurasian-associated Y-chromosome haplogroups are P-M45, and its subclades R and Q. Haplogroup P is inferred to have originated around 44,000 years ago in South or Southeast Asia and is observed with its highest frequency among indigenous Southeast Asian groups, such as the Andamanese or the Jahai people. P is downstream to haplogroup K2b among the Upper Paleolithic Tianyuan man in Northern China, with its sister clade haplogroup MS-P397 being most common among Oceanian populations. Their maternal haplogroup belonged to subclades of haplogroup U commonly found among West Eurasian populations.[17][18][19][20][21]
Formation
The formation of the Ancient North Eurasian/Siberian (ANE/ANS) gene pool occurred during the Upper Paleolithic period: an early West Eurasian population related to Upper Paleolithic Europeans, such as Kostenki-14 and Sungir, migrated via a 'northern route' into Siberia where they encountered and admixed with an Ancient East Eurasian population which arrived earlier during the Initial Upper Paleolithic period via a 'southern route'. The 'East Eurasian' source is most closely related to the ancestry found among the 40,000 year old Tianyuan man of Northern China and other Upper Paleolithic Siberian and East/Southeast Asian groups. In total, the ANE/ANS derive around 50–71% ancestry from an early West Eurasian-affilated source, and 29–50% from an East Eurasian source.[11][23][24][8][9][10][25][26][27][13] On average, the ANE/ANS are usually modeled as roughly 2/3 early West Eurasian and 1/3 early East Eurasian.[28][a]
The Ancient North Eurasian lineage (represented by the Mal'ta and Yana specimens) is taking up an intermediate position between 'Ancient West Eurasians' (represented by a Kostenki-14-like lineage) and 'Ancient East Eurasians' (represented by a Tianyuan-like lineage), consisted with them being the result of a Paleolithic admixture event.[10][9]
Grebenyuk et al. argues that 'Ancient North Eurasians' were "Early Upper Paleolithic tribes of hunters" and linked to similar groups associated with Southern Siberian sites. These communities of Southern Siberian and Central Asian hunters belonged to one of the earliest migration waves of anatomically modern humans into Siberia. The authors summarized that "the initial peopling of Northeastern Asia by the anatomically modern humans could have happened both from West to East and from South to North".[11]
Distribution and expansion patterns
By c. 32kya, populations carrying ANE-related ancestry were probably widely distributed across northeast Eurasia. They may have expanded as far as Alaska and the Yukon, but were forced to abandon high latitude regions following the onset of harsher climatic conditions that came with the Last Glacial Maximum.[30] Gene sequencing of another south-central Siberian people (Afontova Gora-2) dating to approximately 17,000 years ago, revealed similar autosomal genetic signatures to that of Mal'ta boy-1, suggesting that the region was continuously occupied by humans throughout the Last Glacial Maximum.[31]
It is suggested that the ANE ancestry found among modern human populations was largely contributed from a population linked to Afontova Gora (AG2/3), rather than Malta (MA1) or Yana.[32]
ANE-like ancestry was an important genetic contributor to Native Americans, Europeans, Ancient Central Asians, South Asians, and some East Asian groups, in order of significance.[33] Lazaridis et al. (2016:10) note "a cline of ANE ancestry across the east-west extent of Eurasia". A 2016 study found that the global maximum of ANE ancestry occurs in modern-day Kets, Mansi, Native Americans, and Selkups.[6][33]

A deer tooth pendant impregnated with the genetic material of an ANE woman was found in the Denisova Cave, and dated to circa 24,700 years before present. She is closely related to Mal'ta and Afontova Gora specimens, found further east.[34]
The ancient Tianyuan Man and modern East/Southeast Asian populations were found to lack Upper Paleolithic Western Eurasian or ANE-related admixture, suggesting "resistance of those groups to the incoming UP population movements", or alternatively a subsequent reexpansion from a genetically East Asian-like population reservoir.[10] A different but geographically close specimen, known as the Salkhit individual (c. 34,000 BP) from Northern Mongolia was found to display a complex relation to the Yana individuals. While the Yana individuals derived around 33% of their ancestry from a Tianyuan-like source, the Salkhit individual derived around 25% ancestry from the Yana lineage, with the remainder 75% being from the Tianyuan lineage.[28] One study shows high affinities between Tianyuan Man and AR33K, who are both basal to East Asians, and GoyetQ116-1, who is primarily West Eurasian. Compared to Tianyuan Man, AR33K has elevated GoyetQ116-1 ancestry that is 'just at or below the cutoff for statistical significance'. There's also no evidence of the AR33K individual having more Yana-related ancestry than the Tianyuan Man like the Salkhit individual. Similar findings were observed for other Ancient East Asians, suggesting complex population structure before the LGM stadial in northern East Asia, with Yana admixture being mostly confined to Mongolia. The affinities between Tianyuan Man and GoyetQ116-1 can also be explained by shared input from Initial Upper Paleolithic populations that pre-dated the split between European and Asian populations. This tenuously reflects population expansions from Eastern Europe to East Asia.[35][36][37]
Groups partially derived from the Ancient North Eurasians
Native American contribution
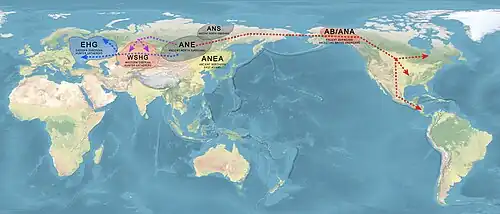
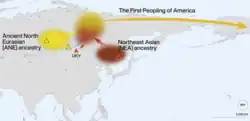
According to Moreno-Mayar et al. 2018 between 14% and 38% of Native American ancestry may originate from gene flow from the Mal'ta–Buret' (ANE) population. This difference is caused by the penetration of posterior "Neo-Siberian" migrations into the Americas, with the lowest percentages of ANE ancestry found in Inuit and Alaskan Natives, as these groups are the result of migrations into the Americas roughly 5,000 years ago.[38] Estimates for ANE ancestry among first wave Native Americans show higher percentages,[39] such as 41% (36-45%) for those belonging to the Andean region in South America.[39] The other ancestry component among Native Americans (the remainder of their ancestry) was of East Asian-like origin, specifically from a lineage that diverged from East Asians c. 30,000 years ago.[31]
According to Jennifer Raff, the Ancient North Eurasian population mixed with a daughter population of ancient East Asians, who they encountered around 25,000 years ago, which led to the emergence of ancestral Native American populations. However, the exact location where the admixture took place is unknown, and the migratory movements that united the two populations are a matter of debate.[40] Vallini et al. 2024 notes that the "position of Native Americans suggests a primarily East Asian ancestry, with a smaller contribution from palaeolithic West Eurasian populations".[41]
One theory supposes that Ancient North Eurasians migrated south to East Asia, or Southern Siberia, where they would have encountered and mixed with ancient East Asians. Genetic evidence from Lake Baikal in Mongolia supports this area as the location where the admixture took place.[42]
However, a third theory, the "Beringian standstill hypothesis", suggests that East Asians instead migrated north to Northeastern Siberia, where they mixed with ANE, and later diverged in Beringia, where distinct Native American lineages formed. This theory is supported by maternal and nuclear DNA evidence.[43] According to Grebenyuk, after 20,000 BP, a branch of Ancient East Asians migrated to Northeastern Siberia, and mixed with descendants of the ANE, leading to the emergence of Ancient Paleo-Siberian and Native American populations in Extreme Northeastern Asia.[44] However, the Beringian standstill hypothesis is not supported by paternal DNA evidence, which may reflect different population histories for paternal and maternal lineages in Native Americans, which is not uncommon and has been observed in other populations.[45]
The descendants of admixture between ANE and ancient East Asians include Ancient Beringian/Ancestral Native American, which are specific archaeogenetic lineages, based on the genome of an infant found at the Upward Sun River site (dubbed USR1), dated to 11,500 years ago.[46] The AB and the Ancestral Native American (ANA) lineage formed about 25,000 years ago, and subsequently diverged from each other, with the AB staying in the Beringian region, while the Ancestral Native Americans populated the Americas. The ANE genetic contribution to late-Paeolithic Ancestral Native Americans (USR1 specimen, dated to 11,500 BP in Alaska, and Clovis specimen, dated to 12,600 BP in Montana) is estimated at 36.8%.[47] There are also the Ancient Paleo-Siberians, populations represented by the Late Upper Paeolithic Lake Baikal Ust'Kyakhta-3 (UKY) 14,050-13,770 BP. They carried 30% ANE ancestry and 70% East Asian ancestry.[3]
Jōmon people
The Jōmon people, the pre-Neolithic population of Japan, primarily descend from an ancient East Asian lineage but have marginally relevant affinities with the Yana Rhinoceros Horn Site specimen, associated with Ancient North Eurasians (or Ancient North Siberians). This indicates gene flow between Ancient North Siberians and the ancestral Jōmon lineage prior to the Jōmon's isolation from other East Eurasians. This gene flow is also associated with the introduction of microblade technology to northern Japan.[48][49] According to Bennett et al. (2024), the Basal Asian-like ancestors of the Jōmon interacted with groups that entered Siberia through a northern migration route, thus explaining the Jōmon's affinities with Ancient North Siberians.[50] Other studies, however, suggest no evidence for direct ANE-related gene flow into the Jōmon, indicating non-demic diffusion. However, the possibility of small non-East Eurasian input is not ruled out.[51][52][53][47][49][54]
Jōmon ancestry is still found among the inhabitants of present-day Japan: most markedly among the Ainu and Ryukyuan peoples, and to a small, but significant degree among the majority of the Japanese population.[55][56][57][58]
Siberian and Asian Holocene populations
Altai hunter-gatherer is the name given to Middle Holocene Siberian hunter-gatherers within the Altai-Sayan region in Southern Siberia. They originated from the admixture of Paleo-Siberian and Ancient North Eurasian groups and show increased affinity towards Native Americans. Bronze Age groups from North and Inner Asia with significant ANE ancestry (e.g. Lake Baikal hunter-gatherers, Okunevo pastoralists) can be successfully modeled with Altai hunter-gatherers as a proximal ANE-derived ancestry source.[59]
West Siberian Hunter-Gatherer (WSHG) is a specific archaeogenetic lineage that was first reported by Narasimhan et al. (2019). It can be modeled as 20% EHG, 73% ANE and 6% Ancient Northeast Asian. Although only represented by three sampled hunter-gatherer individuals from Tyumen Oblast in the Russian Forest Zone east of the Urals dated ca. 5,000 BCE, high-levels of WSHG-like ancestry can be detected in various populations of Central Asia until the Bronze Age. The population of the Botai culture, while probably not directly descended from WSHG, displays a high affinity with the WSHG lineage.[60] The European-Siberian cline defined by Eastern hunter-gatherer-like ancestry stretched from Central Europe to Western Siberia and was already established 10,000 years ago.[61]

Among later Siberian populations, the Neolithic to Early Bronze Age period, Baikal Eneolithic (Baikal_EN) and Baikal Early Bronze Age (Baikal_EBA) derived 6.4% to 20.1% ancestry from ANE, while the rest of their ancestry was derived from Ancient Northeast Asian (ANA) sources. Fofonovo_EN near Lake Baikal were mixture of 12-17% ANE ancestry and 83-87% ANA ancestry.[62]
Present Siberian ancestry found across Eurasia and North America can also be traced to a single gene pool that's best represented by Middle Neolithic Yakutia populations. These populations can be modeled as a mixture of Dzhylinda-1 (71%), which is a mixture of Ancient North Eurasian, Ancient Northern East Asian and Native American, and Early Neolithic West Baikal ancestries (29%), which is Ancient Northern East Asian-rich.[63]
Tarim mummies
A 2021 genetic study on the Tarim mummies found that they were primarily descended from a population represented by the Afontova Gora 3 specimen (AG3), genetically displaying "high affinity" with it.[21] The genetic profile of the Afontova Gora 3 individual represented about 72% of the ancestry of the Tarim mummies, while the remaining 28% of their ancestry was derived from a population represented by the Baikal EBA (Early Bronze Age Northeast Asian Baikal populations).[21]
The Tarim mummies are thus one of the rare Holocene populations who derive most of their ancestry from the Ancient North Eurasians (ANE, specifically the Mal'ta and Afontova Gora populations), despite their distance in time (around 14,000 years).[21] Having survived in a type of "genetic bottleneck" in the Tarim basin where they preserved and perpetuated their ANE ancestry, the Tarim mummies, more than any other ancient populations, can be considered as "the best representatives" of the Ancient North Eurasians among all sampled known Bronze Age populations.[21]
West Asian populations
Mesolithic and Neolithic Iranian populations derived a significant amount of their ancestry from an ANE-like geneflow. ANE ancestry is present among Neolithic Iranians as well as the distantly related Caucasus hunter-gatherers, with the remainder ancestry being largely derived from local or nearby West Eurasian and Basal Eurasian sources.[25][9]
An early Neolithic specimen (Tutkaul1) from Tajikistan was found to be primarily derived from Ancient North Eurasians with some additional Neolithic Iranian-related inputs. The sample is closely related to Afontova Gora 3 (AG3) and Mal’ta 1, as well as to the West Siberian hunter-gatherers (Tyumen and Sosnoviy). While the sample also displays affinity for Eastern hunter-gatherers (EHGs), AG3 was found to be closer to EHGs than Tutkaul1, who instead may be a good proxy for ANE-related ancestry among ancient populations from the Iran and the Turan region.[13]
European populations
Genomic studies also indicate that the ANE component was brought to Western Europe by people related to the Yamnaya culture, long after the Paleolithic.[64][6] Lazaridis et al. (2014) detected ANE ancestry among modern European populations in proportions up to 20%.[65] In ancient European populations, the ANE genetic component is visible in tests of the Yamnaya people[64] but not of Western or Central Europeans predating the Corded Ware culture:[66] ANE ancestry was introduced in the European gene pool with the Eastern hunter-gatherer (EHG) lineage which derived significant ancestry from the ANE, c. 70%, with the remaining ancestry from a group more closely related to, but distinct from, Western hunter-gatherers (WHGs).[33][67][13] It is represented by multiple individuals, such as from Yuzhny Oleny in Karelia, one of Y-haplogroup R1a-M417, dated c. 8.4 kya, the other of Y-haplogroup J, dated c. 7.2 kya; and one individual from Samara, of Y-haplogroup R1b-P297, dated c. 7.6 kya, as well as individuals from Sidelkino and Popovo. After the end of the Last Glacial Maximum, the EHG and WHG lineages merged in Eastern Europe, accounting for early presence of ANE-derived ancestry in Mesolithic Europe. Evidence suggests that as Ancient North Eurasians migrated westward from Eastern Siberia, they absorbed Western Hunter-Gatherers and other West Eurasian populations as well.[68][13]
Villalba-Mouco et al. 2023 confirmed the strong affinity between the Eastern European hunter-gatherers (EHG) to the Ancient North Eurasians, and also found a low affinity to the Tianyuan man, explained by them having received significant amounts of ANE ancestry.[37]
Scandinavian hunter-gatherer (SHG) is represented by several individuals buried at Motala, Sweden ca. 6000 BC. They were descended from Western hunter-gatherers who initially settled Scandinavia from the south, and received later admixture from EHG who entered Scandinavia from the north through the coast of Norway.[69][64][70][71][72]
Western Hunter-Gatherers of the Villabruna cluster also carried the Y-haplogroup R1b, derived from the Ancient North Eurasian haplogroup R*, indicating "an early link between Europe and the western edge of the Steppe Belt of Eurasia."[73]
Western Steppe Herders (WSH) is the name given to a distinct ancestral component that represents descent closely related to the Yamnaya culture of the Pontic–Caspian steppe.[b] This ancestry is often referred to as Yamnaya ancestry or Steppe ancestry, and was formed from EHG and CHG (Caucasus hunter-gatherer) in about equal proportions.[7] The ancient Bronze-age-steppe Yamnaya and Afanasevo cultures were found to have a significant ANE-like component at c. 25–50% via their EHG and CHG ancestry.[64][75]
Phenotype prediction
Genomic studies by Raghavan et al. (2014) and Fu et al. (2016) suggested that Mal'ta boy may have had brown eyes, and relatively dark hair and dark skin,[76][32] while cautioning that this analysis was based on an extremely low coverage of DNA that might not give an accurate prediction of pigmentation.[77] Mathieson, et al. (2018) could not determine if Mal'ta 1 boy had the derived allele associated with blond hair in ANE descendants, as they could obtain no coverage for this SNP.[78]
Anthropologic research

Kozintsev (2020, 2022) argues that the historical Southern Siberian Okunevo population, and other Paleo-Siberians, which derive high amounts of their ancestry from Ancient North Eurasians, as possessing a distinct craniometric phenotype, which he dubbed "Americanoid", which represents the variation of the first humans in Siberia and should not be associated solely with ancient Caucasoids. The Ancient North Eurasians themselves represent a "Boreal" variation of early humans. Craniometric data on ANE-rich remains (such as from Botai), show them to cluster most closely with remains from the Pit-Comb Ware culture in Eastern Europe, and to take up a more "western" position.[79][80]

Zhang et al. (2021) proposed that the 'Western' like features of the earlier Tarim mummies could be attributed to their Ancient North Eurasian ancestry.[21] Previous craniometric analyses on the early Tarim mummies found that they formed their own cluster, and clustered with neither European-related Steppe pastoralists of the Andronovo and Afanasievo cultures, nor with inhabitants of the Western Asian BMAC culture, nor with East Asian populations further east, but displayed an affinity for two specimens from the Harappan site of the Indus Valley Civilisation.[82]
Evolution of blond hair
Blond hair is associated with a single nucleotide polymorphism, the mutated allele rs12821256 of the KITLG gene.[83][84][85][86][87] The earliest known individual with this allele is a female south-central Siberian ANE individual from the Afontova Gora 3 site, which is dated to c. 17,000 before present (the earlier ANE Mal'ta boy lacks the sequence coverage to make this determination).[88] The allele then appears later in ANE-derived Eastern Hunter-Gatherer (EHG) populations at Samara, Motala and Ukraine, circa 10,000 BP, and then in populations with Steppe ancestry.[78] Mathieson, et al. (2018) thus argued that this allele originated in the Ancient North Eurasian population, before spreading to western Eurasia.[78]
Geneticist David Reich said that the KITLG gene for blond hair probably entered continental Europe in a population migration wave from the Eurasian steppe, by a population carrying substantial Ancient North Eurasian ancestry.[89] The gene was also found among the Tarim mummies.[90]
Comparative mythology
Since the term 'Ancient North Eurasian' refers to a genetic bridge of connected mating networks, scholars of comparative mythology have argued that they probably shared myths and beliefs that could be reconstructed via the comparison of stories attested within cultures that were not in contact for millennia and stretched from the Pontic–Caspian steppe to the American continent.[92]
The mytheme of the dog guarding the Otherworld possibly stems from an older Ancient North Eurasian belief, as suggested by similar motifs found in Indo-European, Native American and Siberian mythology. In Siouan, Algonquian, Iroquoian, and in Central and South American beliefs, a fierce guard dog was located in the Milky Way, perceived as the path of souls in the afterlife, and getting past it was a test. The Siberian Chukchi and Tungus believed in a guardian-of-the-afterlife dog and a spirit dog that would absorb the dead man's soul and act as a guide in the afterlife. In Indo-European myths, the figure of the dog is embodied by Cerberus, Sarvarā, and Garmr. In Zoroastrianism, two four-eyed dogs guard the bridge to the afterlife called Chinvat Bridge. Anthony and Brown note that it might be one of the oldest mythemes recoverable through comparative mythology.[92]
A second canid-related series of beliefs, myths and rituals connected dogs with healing rather than death. For instance, Ancient Near Eastern and Turkic-Kipchaq myths are prone to associate dogs with healing and generally categorised dogs as impure. A similar myth-pattern is assumed for the Eneolithic site of Botai in Kazakhstan, dated to 3500 BC, which might represent the dog as absorber of illness and guardian of the household against disease and evil. In Mesopotamia, the goddess Nintinugga, associated with healing, was accompanied or symbolized by dogs. Similar absorbent-puppy healing and sacrifice rituals were practiced in Greece and Italy, among the Hittites, again possibly influenced by Near Eastern traditions.[92]
See also
References
- ^ a b Bednarik 2013.
- ^ a b Lbova 2021.
- ^ a b c Yu et al. 2020.
- ^ a b Yang 2022.
- ^ a b Willerslev & Meltzer 2021.
- ^ a b c Flegontov et al. 2016.
- ^ a b Jeong et al. 2019.
- ^ a b c d Sikora M, Pitulko VV, Sousa VC, Allentoft ME, Vinner L, Rasmussen S, et al. (June 2019). "The population history of northeastern Siberia since the Pleistocene". Nature. 570 (7760): 182–188. Bibcode:2019Natur.570..182S. doi:10.1038/s41586-019-1279-z. hdl:1887/3198847. ISSN 1476-4687. PMC 7617447. PMID 31168093.
- ^ a b c d Vallini L, Zampieri C, Shoaee MJ, Bortolini E, Marciani G, Aneli S, et al. (25 March 2024). "The Persian plateau served as hub for Homo sapiens after the main out of Africa dispersal". Nature Communications. 15 (1): 1882. Bibcode:2024NatCo..15.1882V. doi:10.1038/s41467-024-46161-7. ISSN 2041-1723. PMC 10963722. PMID 38528002.
- ^ a b c d e Vallini et al. 2022
- ^ a b c Grebenyuk et al. 2022.
- ^ Raff J (8 February 2022). Origin: A Genetic History of the Americas. Grand Central Publishing. ISBN 978-1-5387-4970-8.
- ^ a b c d e Posth et al. 2023.
- ^ Reich D (2018). "4. Humanity's Ghosts". Who We Are and How We Got Here: Ancient DNA and the New Science of the Human Past. Oxford University Press. p. 96. ISBN 978-0-19-882125-0.
- ^ Olsen BA, Olander T, Kristiansen K (23 August 2019). Tracing the Indo-Europeans: New evidence from archaeology and historical linguistics. Oxbow Books. ISBN 978-1-78925-273-6.
- ^ "Hall 11 panorama". State Hermitage Museum.
- ^ "The Mal'ta - Buret' venuses and culture in Siberia". donsmaps.com.
- ^ Estes R (12 September 2020). "Y DNA Haplogroup P Gets a Brand-New Root – Plus Some Branches". DNAeXplained - Genetic Genealogy. Retrieved 27 June 2024.
- ^ "P YTree". www.yfull.com. Retrieved 27 July 2024.
formed 44300 ybp, TMRCA 41500 ybp
- ^ "Allen Ancient DNA Resource (AADR): Downloadable genotypes of present-day and ancient DNA data". reich.hms.harvard.edu. David Reich Lab. Retrieved 21 January 2024.
- ^ a b c d e f g h Zhang et al. 2021.
- ^ Allentoft et al. 2024, pp. 301–311, Supplementary Information: Fig. S3d.16. Admixture graph of deep Eurasian lineages.
- ^ Zhang X, Ji X, Li C, Yang T, Huang J, Zhao Y, et al. (25 July 2022). "A Late Pleistocene human genome from Southwest China". Current Biology. 32 (14): 3095–3109.e5. doi:10.1016/j.cub.2022.06.016. PMID 35839766.
- ^ 张明, 平婉菁, Yang MA, 付巧妹 (2023). "古基因组揭示史前欧亚大陆现代人复杂遗传历史" [Ancient genomes reveal complex genetic history of modern humans in prehistoric Eurasia]. 人类学学报 [Journal of Anthropology] (in Chinese). 42 (3): 412–421. doi:10.16359/j.1000-3193/aas.2023.0010.
- ^ a b Allentoft ME, Sikora M, Refoyo-Martínez A, Irving-Pease EK, Fischer A, Barrie W, et al. (January 2024). "Population genomics of post-glacial western Eurasia". Nature. 625 (7994): 301–311. Bibcode:2024Natur.625..301A. doi:10.1038/s41586-023-06865-0. ISSN 1476-4687. PMC 10781627. PMID 38200295.
- ^ Yang et al. 2020.
- ^ Yang MA, Gao X, Theunert C, Tong H, Aximu-Petri A, Nickel B, et al. (23 October 2017). "40,000-Year-Old Individual from Asia Provides Insight into Early Population Structure in Eurasia". Current Biology. 27 (20): 3202–3208.e9. Bibcode:2017CBio...27E3202Y. doi:10.1016/j.cub.2017.09.030. ISSN 0960-9822. PMC 6592271. PMID 29033327.
- ^ a b Massilani D, Skov L, Hajdinjak M, Gunchinsuren B, Tseveendorj D, Yi S, et al. (30 October 2020). "Denisovan ancestry and population history of early East Asians". Science. 370 (6516): 579–583. doi:10.1126/science.abc1166. PMID 33122380.
- ^ "Figurine of a woman with the "Spanish rose" hairdo". Art of Mal'ta. Novosibirsk State University.
- ^ Maritime Prehistory of Northeast Asia. The Archaeology of Asia-Pacific Navigation. Vol. 6. 2022. doi:10.1007/978-981-19-1118-7. ISBN 978-981-19-1117-0.
- ^ a b Raghavan et al. 2013.
- ^ a b Fu et al. 2016.
- ^ a b c Lazaridis et al. 2016.
- ^ a b Essel E (3 May 2023). "Ancient human DNA recovered from a Palaeolithic pendant". Nature. 618 (7964): 328–332. Bibcode:2023Natur.618..328E. doi:10.1038/s41586-023-06035-2. PMC 10247382. PMID 37138083.
- ^ Mao X, Zhang H, Qiao S, Liu Y, Chang F, Xie P, et al. (June 2021). "The deep population history of northern East Asia from the Late Pleistocene to the Holocene". Cell. 184 (12): 3256–3266.e13. doi:10.1016/j.cell.2021.04.040. PMID 34048699.
- ^ Hajdinjak M, Mafessoni F, Skov L, Vernot B, Hübner A, Fu Q, et al. (8 April 2021). "Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry". Nature. 592 (7853): 253–257. Bibcode:2021Natur.592..253H. doi:10.1038/s41586-021-03335-3. PMC 8026394. PMID 33828320.
- ^ a b Villalba-Mouco et al. 2023.
- ^ Moreno-Mayar et al. 2018.
- ^ a b Wong et al. 2017.
- ^ Raff 2022, p. 188.
- ^ Vallini et al. 2024, p. 1882, "Similarly, Mal’ta and Yana fall in an intermediate position between the two axes, the result of a palaeolithic admixture between EEC and WEC groups18.".
- ^ Raff 2022, p. 188
- ^ Raff 2022, pp. 188–189
- ^ Grebenyuk et al. 2022, p. 96.
- ^ Hoffecker JF, Elias SA, O'Rourke DH, Scott GR, Bigelow NH (4 March 2016). "Beringia and the global dispersal of modern humans". Evolutionary Anthropology: Issues, News, and Reviews. 25 (2): 64–78. doi:10.1002/evan.21478. PMID 27061035.
- ^ Moreno-Mayar et al. 2018.
- ^ a b Gakuhari T, Nakagome S, Rasmussen S, Allentoft ME (25 August 2020). "Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations". Communications Biology. 3 (1): Fig.1 A, C. doi:10.1038/s42003-020-01162-2. ISSN 2399-3642. PMC 7447786. PMID 32843717.
- ^ Cooke NP, Mattiangeli V, Cassidy LM, Okazaki K, Stokes CA, Onbe S, et al. (2021). "Ancient genomics reveals tripartite origins of Japanese populations". Science Advances. 7 (38): eabh2419. Bibcode:2021SciA....7.2419C. doi:10.1126/sciadv.abh2419. PMC 8448447. PMID 34533991.
However, we note no dilution of Jomon ancestry in the Japanese population (15.0 ± 3.8%), relative to the Kofun individuals (13.1 ± 3.5%) (fig. S22).
- ^ a b Osada & Kawai 2021.
- ^ Bennett AE, Liu Y, Fu Q (2025). Reconstructing the Human Population History of East Asia through Ancient Genomics. doi:10.1017/9781009246675. ISBN 978-1-009-24667-5.
- ^ Kanzawa-Kiriyama et al. 2017.
- ^ Kanzawa-Kiriyama H, Jinam TA, Kawai Y, Sato T, Hosomichi K, Tajima A, et al. (2019). "Late Jomon male and female genome sequences from the Funadomari site in Hokkaido, Japan". Anthropological Science. 127 (2): 83–108. doi:10.1537/ase.190415.
Next, we assessed the possibility of gene flow between non-East Asians and the ancestor of Funadomari Jomons to explain the deep divergence. f4 (African, non-East Asian; East Asian, F23) showed that no modern populations, except for Sardinian, Relli, and Aboriginal Australian, who had small positive f4 values (but without statistical significance), supported the gene flow. The 24000 year old MA1 also did not support the gene flow.
- ^ de Boer E, Yang MA, Kawagoe A, Barnes GL (2020). "Japan considered from the hypothesis of farmer/language spread". Evolutionary Human Sciences. 2 e13. doi:10.1017/ehs.2020.7. PMC 10427481. PMID 37588377.
Ancient northern Siberian ancestry prevalent during the Palaeolithic notable for both its closer relationship with European-related rather than Asian-related ancestry and its impact on Native American ancestry is not found in mainland East Asians or the Jōmon, which emphasizes that the connections are specific to coastal mainland East Asians and the Jōmon.
- ^ Abood S, Oota H (14 February 2025). "Human dispersal into East Eurasia: ancient genome insights and the need for research on physiological adaptations". Journal of Physiological Anthropology. 44 (1): 5. doi:10.1186/s40101-024-00382-3. PMC 11829451. PMID 39953642.
However, the fact that no evidence of gene flow from Mal'ta was detected in the genome of the Ikawazu Jomon individual was a novel discovery...To date, the conclusion remains unchanged. However, not all Jomon genome sequences analyzed so far have undergone the type of southern versus northern route-focused analysis presented in Gakuhari et al. (2020). Conducting further analysis on this issue remains a task for the future.
- ^ Watanabe Y, Ohashi J (2022). Modern Japanese ancestry-derived variants revealed the formation process of the current Japanese regional gradations (Preprint). doi:10.1101/2020.12.07.414037.
- ^ "日本人のルーツ「縄文人」とは?". app.zene360.com. Retrieved 6 May 2025.
- ^ "縄文人と渡来人の混血史から日本列島人の地域的多様性の起源を探る". 東京大学 大学院理学系研究科・理学部 (in Japanese). Retrieved 6 May 2025.
- ^ Yamamoto K, Namba S, Sonehara K, Suzuki K, Sakaue S, Cooke NP, et al. (12 November 2024). "Genetic legacy of ancient hunter-gatherer Jomon in Japanese populations". Nature Communications. 15 (1): 9780. Bibcode:2024NatCo..15.9780Y. doi:10.1038/s41467-024-54052-0. ISSN 2041-1723. PMC 11558008. PMID 39532881.
In the whole BBJ dataset, the proportions of the three distinct ancestral components closely align with those reported in the previous study (Jomon: 12.4%, Northeast Asia: 21.2%, and East Asia: 66.4%)14. However, Jomon ancestry exhibits regional variation, ranging from 9.8% in Kinki to 26.1% in Okinawa (Fig. 2a). Within the Ryukyu Islands, there is an elevated level of Jomon ancestry, with the highest proportion observed on Yoron Island (Fig. 2b). Jomon ancestry is even higher in one of the genetically-defined populations, Hokkaido_sub (31.6%, Fig. 2c and Supplementary Data 3), which primarily includes a subset of individuals from Hokkaido.
- ^ Wang K, Yu H, Radzevičiūtė R, Kiryushin YF, Tishkin AA, Frolov YV, et al. (12 January 2023). "Middle Holocene Siberian genomes reveal highly connected gene pools throughout North Asia". Current Biology. 33 (3): 423–433. Bibcode:2023CBio...33E.423W. doi:10.1016/j.cub.2022.11.062. ISSN 0960-9822. PMID 36638796.
- ^ Narasimhan 2019.
- ^ Mattila TM, Svensson EM, Juras A, Günther T, Kashuba N, Ala-Hulkko T, et al. (9 August 2023). "Genetic continuity, isolation, and gene flow in Stone Age Central and Eastern Europe". Communications Biology. 6 (1): 793. doi:10.1038/s42003-023-05131-3. hdl:10852/104260. ISSN 2399-3642. PMC 10412644. PMID 37558731.
- ^ Jeong et al. 2020.
- ^ Gill H, Lee J, Jeong C (2023). Reconstructing the genetic relationship between ancient and present-day Siberian populations (Preprint). doi:10.1101/2023.08.21.554074.
- ^ a b c d Haak et al. 2015.
- ^ Lazaridis I, Patterson N, Mittnik A (September 2014). "Ancient human genomes suggest three ancestral populations for present-day Europeans". Nature. 513 (7518): 409–413. arXiv:1312.6639. Bibcode:2014Natur.513..409L. doi:10.1038/nature13673. ISSN 1476-4687. PMC 4170574. PMID 25230663.
- ^ Lazaridis, Patterson & Mittnik 2014.
- ^ Wang 2019.
- ^ Mathieson et al. 2018.
- ^ Lazaridis et al. 2014.
- ^ Mathieson 2015.
- ^ Günther et al. 2018.
- ^ Mittnik 2018.
- ^ Gabidullina LR, Dzhaubermezov MA, Ekomasova NV, Sufyanova ZR, Khusnutdinova EK (2023). "Genetic History of Eurasia Before the Common Era". Opera Medica et Physiologica. 10 (3): 95–117. ISSN 2500-2295.
- ^ Jeong et al. 2018.
- ^ Kozintsev AG (31 December 2020). "The Origin of the Okunev Population, Southern Siberia: The Evidence of Physical Anthropology and Genetics". Archaeology, Ethnology & Anthropology of Eurasia. 48 (4): 135–145. doi:10.17746/1563-0110.2020.48.4.135-145.
- ^ Raghavan & Skoglund et al. 2014.
- ^ Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, et al. (2 January 2014). "Upper Palaeolithic Siberian genome reveals dual ancestry of NativeAmericans". Nature. 505 (7481): 87–91. Bibcode:2014Natur.505...87R. doi:10.1038/nature12736. ISSN 0028-0836. PMC 4105016. PMID 24256729.
- ^ a b c Mathieson et al. 2018 "Supplementary Information page 52
- ^ Kozintsev AG (31 December 2020). "The Origin of the Okunev Population, Southern Siberia: The Evidence of Physical Anthropology and Genetics". Archaeology, Ethnology & Anthropology of Eurasia. 48 (4): 135–145. doi:10.17746/1563-0110.2020.48.4.135-145.
- ^ Kozintsev AG (4 January 2022). "Patterns in the Population History of Northern Eurasia from the Mesolithic to the Early Bronze Age, Based on Craniometry and Genetics". Archaeology, Ethnology & Anthropology of Eurasia. 49 (4): 140–151. doi:10.17746/1563-0110.2021.49.4.140-151.
Judging by the huge distribution area of ANE, it was a legacy of early Homo sapiens, members of the Boreal meta-population (Biasutti, 1941: 275; Kozintsev, 2013, 2014) [...] The ancestor of ANE was the ANS (Ancient North Siberian) autosomal component, represented in a male from the Upper Paleolithic Yana site, dating to 31.6 ka BP (Sikora et al., 2019). ANS is thought to have originated among West Eurasians soon after their divergence from East Eurasians about 43 ka BP. [...] The sample that includes the few Botai crania (No. 24) takes a more "western" position (Fig. 1), forming a pair with the Pit-Comb Ware group of European Russia, and this is supported by archaeological data suggesting that these cultures are related (Mosin, 2003: 97–98)
- ^ Zhang 2021: "The Tarim mummies are among only a few known Holocene populations that derive the majority of their ancestry from Pleistocene ANE groups, who once made up the huntergatherer populations of southern Siberia, and which are represented by individual genomes from the archaeological sites of Mal'ta (MA-1)29 and Afontova Gora (AG3). (...) The Tarim mummies are currently the best representative of the pre-pastoralist ANE-related population that once inhabited Central Asia and southern Siberia (Extended Data Fig. 2A), even though Tarim_EMBA1 postdates these populations in time."
- ^ Hemphill BE, Mallory J (July 2004). "Horse-mounted invaders from the Russo-Kazakh steppe or agricultural colonists from western Central Asia? A craniometric investigation of the Bronze Age settlement of Xinjiang". American Journal of Physical Anthropology. 124 (3): 199–222. doi:10.1002/ajpa.10354. PMID 15197817.
- ^ Carlberg & Hanel 2020.
- ^ Hartz P. "KIT LIGAND; KITLG". Online Mendelian Inheritance in Man.
- ^ Khan R. "KITLG makes you white skinned?". Discover Magazine.
- ^ "P21583". Uniprot.
- ^ Guenther et al. 2014.
- ^ Evans G (2019). Skin Deep: Dispelling the Science of Race (1 ed.). Simon and Schuster. p. 139. ISBN 978-1-78607-623-6.
- ^ Reich D (2018). Who We are and How We Got Here: Ancient DNA and the New Science of the Human Past. Oxford University Press. ISBN 978-0-19-882125-0.
- ^ Zhang 2021.
- ^ "The Mal'ta - Buret' venuses and culture in Siberia". donsmaps.com.
- ^ a b c Anthony & Brown 2019, pp. 104–106.
Notes
- ^ Sikora et al. (2019) model the ANE/ANS individuals as 22–29% East Eurasian and the remainder West Eurasian. Massilani et al. (2020): "In agreement with previous results (10), the Yana individuals are estimated to have about one-third of their ancestry from early East Eurasians and the remaining two-thirds from the early West Eurasians." Yang et al. (2020) modeled the ANS/ANE as c. 32–38% East Eurasian and c. 62–68% West Eurasian. Vallini et al. (2022) model Yana as c. 50% West Eurasian and c. 50% East Eurasian. Vallini et al. (2024) modeled ANS (Yana) as around 47% East Eurasian and 53% West Eurasian, while ANE (Malta) was modeled as roughly 33% East Eurasian and 67% West Eurasian. Allentoft et al. (2024) modeled both ANS/ANE as around 35% East Eurasian and 65% West Eurasian.
- ^ "Recent paleogenomic studies have shown that migrations of Western steppe herders (WSH) beginning in the Eneolithic (ca. 3300–2700 BCE) profoundly transformed the genes and cultures of Europe and central Asia... The migration of these Western steppe herders (WSH), with the Yamnaya horizon (ca. 3300–2700 BCE) as their earliest representative, contributed not only to the European Corded Ware culture (ca. 2500–2200 BCE) but also to steppe cultures located between the Caspian Sea and the Altai-Sayan mountain region, such as the Afanasievo (ca. 3300–2500 BCE) and later Sintashta (2100–1800 BCE) and Andronovo (1800–1300 BCE) cultures."[74]
Bibliography
- Anthony DW, Brown DR (2019). "Late Bronze Age midwinter dog sacrifices and warrior initiations at Krasnosamarskoe, Russia". In Olsen BA, Olander T, Kristiansen K (eds.). Tracing the Indo-Europeans: New evidence from archaeology and historical linguistics. Oxbow Books. ISBN 978-1-78925-273-6.
- Balter M (25 October 2013). "Ancient DNA Links Native Americans With Europe". Science. 342 (6157): 409–410. Bibcode:2013Sci...342..409B. doi:10.1126/science.342.6157.409. PMID 24159019.
- Bednarik R (19 June 2013). "Pleistocene Palaeoart of Asia". Arts. 2 (2): 46–76. doi:10.3390/arts2020046.
- Dolitsky AB, Ackerman RE, Aigner JS, Bryan AL, Dennell R, Guthrie RD, et al. (June 1985). "Siberian Paleolithic Archaeology: Approaches and Analytic Methods [and Comments and Replies]". Current Anthropology. 26 (3): 361–378. doi:10.1086/203280.
- Carlberg C, Hanel A (2020). "Skin colour and vitamin D: An update". Experimental Dermatology. 29 (9): 864–875. doi:10.1111/exd.14142. PMID 32621306.
- Flegontov P, Changmai P, Zidkova A, Logacheva MD, Altınışık NE, Flegontova O, et al. (11 February 2016). "Genomic study of the Ket: a Paleo-Eskimo-related ethnic group with significant ancient North Eurasian ancestry". Scientific Reports. 6 (1): 20768. arXiv:1508.03097. Bibcode:2016NatSR...620768F. doi:10.1038/srep20768. PMC 4750364. PMID 26865217.
- Fu Q, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes D, et al. (9 June 2016). "The genetic history of Ice Age Europe". Nature. 534 (7606): 200–205. Bibcode:2016Natur.534..200F. doi:10.1038/nature17993. PMC 4943878. PMID 27135931.
- Günther T, Malmström H, Svensson EM, Omrak A, Sánchez-Quinto F, Kılınç GM, et al. (9 January 2018). "Population genomics of Mesolithic Scandinavia: Investigating early postglacial migration routes and high-latitude adaptation". PLOS Biology. 16 (1): e2003703. doi:10.1371/journal.pbio.2003703. PMC 5760011. PMID 29315301.
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, et al. (2 March 2015). "Massive migration from the steppe was a source for Indo-European languages in Europe". Nature. 522 (7555): 207–211. arXiv:1502.02783. Bibcode:2015Natur.522..207H. doi:10.1038/nature14317. PMC 5048219. PMID 25731166.
- Guenther CA, Tasic B, Luo L, Bedell MA, Kingsley DM (July 2014). "A molecular basis for classic blond hair color in Europeans". Nature Genetics. 46 (7): 748–752. doi:10.1038/ng.2991. PMC 4704868. PMID 24880339.
- Grebenyuk PS, Fedorchenko AY, Dyakonov VM, Lebedintsev AI, Malyarchuk BA (2022). "Ancient Cultures and Migrations in Northeastern Siberia". Humans in the Siberian Landscapes. Springer Geography. pp. 89–133. doi:10.1007/978-3-030-90061-8_4. ISBN 978-3-030-90060-1.
- Jeong C, Wilkin S, Amgalantugs T, Bouwman AS, Taylor WT, Hagan RW, et al. (27 November 2018). "Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe". Proceedings of the National Academy of Sciences of the United States of America. 115 (48): E11248 – E11255. Bibcode:2018PNAS..11511248J. doi:10.1073/pnas.1813608115. ISSN 0027-8424. PMC 6275519. PMID 30397125.
- Jeong C, Balanovsky O, Lukianova E, Kahbatkyzy N, Flegontov P, Zaporozhchenko V, et al. (June 2019). "The genetic history of admixture across inner Eurasia". Nature Ecology & Evolution. 3 (6): 966–976. Bibcode:2019NatEE...3..966J. doi:10.1038/s41559-019-0878-2. PMC 6542712. PMID 31036896.
- Jeong C, Wang K, Wilkin S, Taylor WT, Miller BK, Ulziibayar S, et al. (26 March 2020). "A dynamic 6,000-year genetic history of Eurasia's Eastern Steppe". Cell. 183 (4): 890–904.e29. bioRxiv 10.1101/2020.03.25.008078. doi:10.1016/j.cell.2020.10.015. hdl:21.11116/0000-0007-77BF-D. PMC 7664836. PMID 33157037.
- Kanzawa-Kiriyama H, Kryukov K, Jinam TA, Hosomichi K, Saso A, Suwa G, et al. (February 2017). "A partial nuclear genome of the Jomons who lived 3000 years ago in Fukushima, Japan". Journal of Human Genetics. 62 (2): 213–221. doi:10.1038/jhg.2016.110. PMC 5285490. PMID 27581845.
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, et al. (25 July 2016). "Genomic insights into the origin of farming in the ancient Near East". Nature. 536 (7617): 419–424. Bibcode:2016Natur.536..419L. doi:10.1038/nature19310. PMC 5003663. PMID 27459054.
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, et al. (17 September 2014). "Ancient human genomes suggest three ancestral populations for present-day Europeans". Nature. 513 (7518): 409–413. arXiv:1312.6639. Bibcode:2014Natur.513..409L. doi:10.1038/nature13673. hdl:11336/30563. PMC 4170574. PMID 25230663.
{{cite journal}}: CS1 maint: overridden setting (link) - Lbova L (2021). "The Siberian Paleolithic site of Mal'ta: a unique source for the study of childhood archaeology". Evolutionary Human Sciences. 3 e9. doi:10.1017/ehs.2021.5. PMC 10427291. PMID 37588521.
- Maier R, Flegontov P, Flegontova O, Isildak U, Changmai P, Reich D (2023). "On the limits of fitting complex models of population history to f-statistics". eLife. 12 e85492. doi:10.7554/eLife.85492. PMC 10310323. PMID 37057893.
- Diyendo Massilani, Laurits Skov, Mateja Hajdinjak, Byambaa Gunchinsuren, Damdinsuren Tseveendorj, Seonbok Yi, et al. (2020). "Denisovan ancestry and population history of early East Asians". Science. 370 (6516): 579–583. doi:10.1126/science.abc1166. PMID 33122380.
- Mathieson I, Alpaslan-Roodenberg S, Posth C, Szécsényi-Nagy A, Rohland N, Mallick S, et al. (21 February 2018). "The genomic history of southeastern Europe". Nature. 555 (7695): 197–203. Bibcode:2018Natur.555..197M. doi:10.1038/nature25778. hdl:11392/2418484. PMC 6091220. PMID 29466330.
{{cite journal}}: CS1 maint: overridden setting (link) - Mathieson I (23 November 2015). "Genome-wide patterns of selection in 230 ancient Eurasians". Nature. 528 (7583). Nature Research: 499–503. Bibcode:2015Natur.528..499M. doi:10.1038/nature16152. PMC 4918750. PMID 26595274.
- Mittnik A (30 January 2018). "The genetic prehistory of the Baltic Sea region". Nature Communications. 9 (1) 442. Nature Research. Bibcode:2018NatCo...9..442M. doi:10.1038/s41467-018-02825-9. PMC 5789860. PMID 29382937.
- Moreno-Mayar JV, Vinner L, de Barros Damgaard P, de la Fuente C, Chan J, Spence JP, et al. (7 December 2018). "Early human dispersals within the Americas". Science. 362 (6419): eaav2621. Bibcode:2018Sci...362.2621M. doi:10.1126/science.aav2621. PMID 30409807.
- Moreno-Mayar JV, Potter BA, Vinner L, Steinrücken M, Rasmussen S, Terhorst J, et al. (January 2018). "Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans" (PDF). Nature. 553 (7687): 203–207. Bibcode:2018Natur.553..203M. doi:10.1038/nature25173. PMID 29323294.
- Narasimhan VM (6 September 2019). "The formation of human populations in South and Central Asia". Science. 365 (6457) eaat7487. American Association for the Advancement of Science. bioRxiv 10.1101/292581. doi:10.1126/science.aat7487. PMC 6822619. PMID 31488661.
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, et al. (20 November 2013). "Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans". Nature. 505 (7481): 87–91. Bibcode:2014Natur.505...87R. doi:10.1038/nature12736. PMC 4105016. PMID 24256729.
- Posth C, Yu H, Ghalichi A, Rougier H, Crevecoeur I, Huang Y, et al. (March 2023). "Palaeogenomics of Upper Palaeolithic to Neolithic European hunter-gatherers". Nature. 615 (7950): 117–126. Bibcode:2023Natur.615..117P. doi:10.1038/s41586-023-05726-0. PMC 9977688. PMID 36859578.
- Raghavan M, Skoglund P, Graf KE (2014). "Upper Palaeolithic Siberian Genome Reveals Dual Ancestry of Native Americans". Nature. 505 (7481): 87–91. Bibcode:2014Natur.505...87R. doi:10.1038/nature12736. PMC 4105016. PMID 24256729.
- Osada N, Kawai Y (2021). "Exploring models of human migration to the Japanese archipelago using genome-wide genetic data". Anthropological Science. 129 (1): 45–58. doi:10.1537/ase.201215.
- Vallini L, Marciani G, Aneli S, Bortolini E, Benazzi S, Pievani T, et al. (2022). "Genetics and Material Culture Support Repeated Expansions into Paleolithic Eurasia from a Population Hub Out of Africa". Genome Biology and Evolution. 14 (4) evac045. doi:10.1093/gbe/evac045. PMC 9021735. PMID 35445261.
- Villalba-Mouco V, van de Loosdrecht MS, Rohrlach AB, Fewlass H, Talamo S, Yu H, et al. (March 2023). "A 23,000-year-old southern Iberian individual links human groups that lived in Western Europe before and after the Last Glacial Maximum". Nature Ecology & Evolution. 7 (4): 597–609. Bibcode:2023NatEE...7..597V. doi:10.1038/s41559-023-01987-0. hdl:10230/57143. PMC 10089921. PMID 36859553.
- Wang CC (4 February 2019). "Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions Eurasia". Nature Communications. 10 (1). Nature Research: 590. Bibcode:2019NatCo..10..590W. doi:10.1038/s41467-018-08220-8. PMC 6360191. PMID 30713341.
- Willerslev E, Meltzer DJ (June 2021). "Peopling of the Americas as inferred from ancient genomics". Nature. 594 (7863): 356–364. Bibcode:2021Natur.594..356W. doi:10.1038/s41586-021-03499-y. PMID 34135521.
- Wong EH, Khrunin A, Nichols L, Pushkarev D, Khokhrin D, Verbenko D, et al. (January 2017). "Reconstructing genetic history of Siberian and Northeastern European populations". Genome Research. 27 (1): 1–14. doi:10.1101/gr.202945.115. PMC 5204334. PMID 27965293.
- Yang MA, Fan X, Sun B, Chen C, Lang J, Ko YC, et al. (17 July 2020). "Ancient DNA indicates human population shifts and admixture in northern and southern China". Science. 369 (6501): 282–288. Bibcode:2020Sci...369..282Y. doi:10.1126/science.aba0909. PMID 32409524.
- Yang MA (6 January 2022). "A genetic history of migration, diversification, and admixture in Asia". Human Population Genetics and Genomics. 2 (1) 0001: 1–32. doi:10.47248/hpgg2202010001.
- Yu H, Spyrou MA, Karapetian M, Shnaider S, Radzevičiūtė R, Nägele K, et al. (June 2020). "Paleolithic to Bronze Age Siberians Reveal Connections with First Americans and across Eurasia". Cell. 181 (6): 1232–1245.e20. doi:10.1016/j.cell.2020.04.037. PMID 32437661.
- Zhang F, Ning C, Scott A, Fu Q, Bjørn R, Li W, et al. (11 November 2021). "The genomic origins of the Bronze Age Tarim Basin mummies". Nature. 599 (7884): 256–261. Bibcode:2021Natur.599..256Z. doi:10.1038/s41586-021-04052-7. PMC 8580821. PMID 34707286.
.jpg)
.png)

.png)